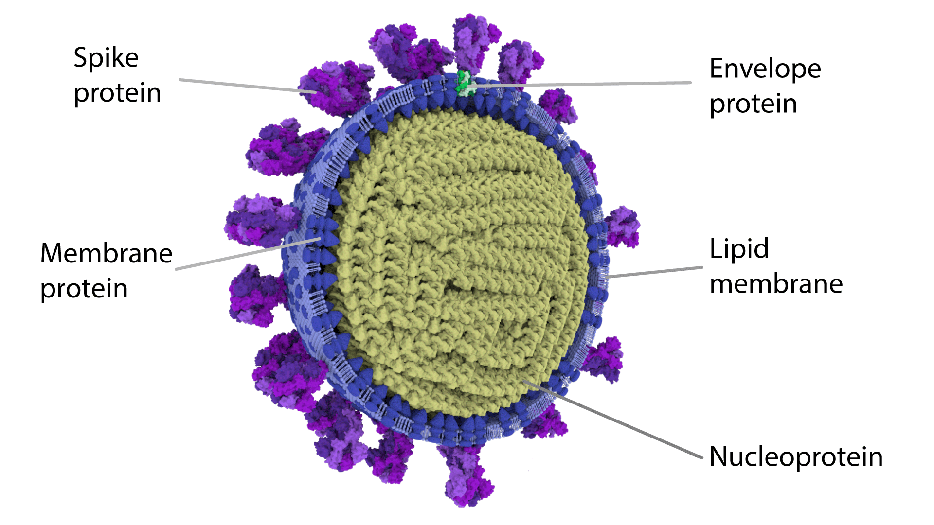
Image showing the most detailed 3D half view of SARS CoV-2 virion.
In a recent preliminary research (not peer-reviewed yet), two teams of researchers found a second protein that SARS-CoV-2 uses to enter human cells.
The spike protein (S) of the SARS-CoV-2 is all responsible for host cell attachment and penetration. Prior to its entry and infection of cells, by S protein on its surface, the SARS-CoV-2 virus binds to angiotensin-converting enzyme 2 (ACE2). ACE2 acts as a cellular doorway – a receptor – for the virus that causes COVID-19.
Research on the development and design of antiviral drug/ vaccine of SARS-CoV-2 are primarily focused on blocking/ treating the virus from easily entering into the host cell which is responsible for the infection. They are mainly focused on S-protein structure and its connection to ACE2 receptors.
The viral glycoprotein (carbohydrate groups of proteins attached to the polypeptide chain) is synthesized as a precursor in infected cells and, to be active, they must be cleaved to two associated polypeptides: S1 and S2.
Teams of researchers found in addition to engaging the known receptor ACE2 S1, one of the two associated polypeptides, can bind to neuropilin-1 (NRP1) through the canonical C-end rule (CendR) mechanism.
The research work was published in the journal bioRxiv.
In the S protein of SARS-CoV-2, there is the presence of a polybasic sequence motif, called Arg-Arg-Ala-Arg (RRAR), but in SARS-CoV, this polybasic sequence is all absent. This sequence motif conforms to the C-end rule, which means that the C-terminal sequence may allow the protein to associate with cell surface neuropilin-1 and neuropilin-2 (NRP2) receptors.
The team engineered a HEK293T cell line to stably express SARS-CoV-2 S protein and explore the possible association of the SARS-CoV-2 S1 protein with neuropilins.
Human embryonic kidney 293 cells (HEK293 cell) are a specific cell line originally derived from human embryonic kidney cells grown in tissue culture and HEK 293T is one of the variants of HEK293 cell.
Earlier, scientists confirmed that the SARS-CoV-2, virus which causes COVID-19, is mutating and even identified 6,822 mutations of SARS-CoV-2. But their study suggests us not to panic.
Tagging NPR1 at the C-terminus with GFP (NRP1-GFP) researchers found that NRP1-GFP was associated with a number of proteins some of which could be S1-processed versions of the S protein.
Further analysis showed that the SARS-CoV-2 S-protein binds to cell surface receptor NRP1 via the S1 CendR motif generated by the furin cleavage of S1/S2. This basically signifies that a host cell surface neuropilin-1 (NRP1) is also responsible for the SARS-CoV-2 infection.
Also, in a second study published in bioRxiv (have not been peer-reviewed yet) observed that the cellular receptor neuropilin-1 (NRP1) significantly potentiates SARS-CoV-2 infectivity.
Researchers from the Technical University of Munich, Germany, and colleagues performed experiments in mice and determined whether NRPs could provide a pathway for virus entry into the brain through the olfactory system. The team explored SARS-CoV-2 isolated from COVID-19 patients from the Helsinki University Hospital.
They practically observed that NRP1 assists the entry of virus-sized particles into the central nervous system.
RNA viruses such as SARS-CoV-2 have a remarkable ability to adapt to their host environment by generating adaptive mutations in a short period.
Though more verifications are needed to be confirmed, researchers are hopeful that this preliminary study will be helpful to conduct more research on this topic and will assist in developing and designing the antiviral drug/vaccine.